"Volume Studio" - Visualization of Medical Data
Volume Studio is a software package developed by the University of Paderborn to support volume rendering of medical data, such as CT and PET data.
Introduction
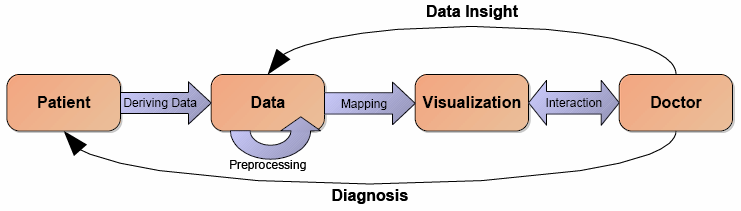
From the standpoint of Computer Science, visualization of medical data means mapping data generated from a patient by means of medical measuring devices into a picture (or collection of pictures) to be interpreted by a medical doctor. There are several stages inherent in the process as seen in Figure 1.
Between 2005 and 2015, a tight collaboration existed with the Institute for Radiology, Nuclear Medicine and Molecular Imaging at the Heart- and Diabetes Center North-Rhine Westphalia (Prof. Dr. med. W. Burchert, Dr. med. E. Fricke, Dipl.-Ing. R. Weise, Dr. H. Fricke) in exploring multi modality imaging and visualization of cardiac disease. Together we worked on improving non-invasive diagnoses support (based on CT, MRI, PET data) to minimize risks for the patient when planning a therapy.
Transfer Functions and more
Our research focused on comparing the value of different transfer functions, on movement-corrected reconstruction of PET listmode data and segmentation of the coronary tree for CT angiography (Preprocessing step in Figure 1). For the mapping stage, we developed automatic classification methods to classify CT and PET data. Fusion of various imaging modes is an important part of visualization. All algorithms are developed on modern graphics boards and use the GPU extensively.
Volume Rendering with Volume Studio
Volume Studio is a software package developed by our research group at the University of Paderborn to support volume rendering of medical data, such as CT and PET data. Our main interest is in new/advanced methods that physicians can actually apply in their daily work. Therefore, direct volume rendering is a powerful technique for multi-modality visualization of CT and PET data as it has the potential to show the three dimensional structure of a feature of interest, rather than just a small part of the data by a cutting plane. It helps the viewer to get a better insight into the relative 3D positions of the object components and makes it easier to detect and understand complex phenomena like coronary stenosis for diagnostic and operation planning. The latest advances in consumer graphic cards has made this approach even more attractive, as high quality interactive volume rendering is now possible on common low cost hardware.
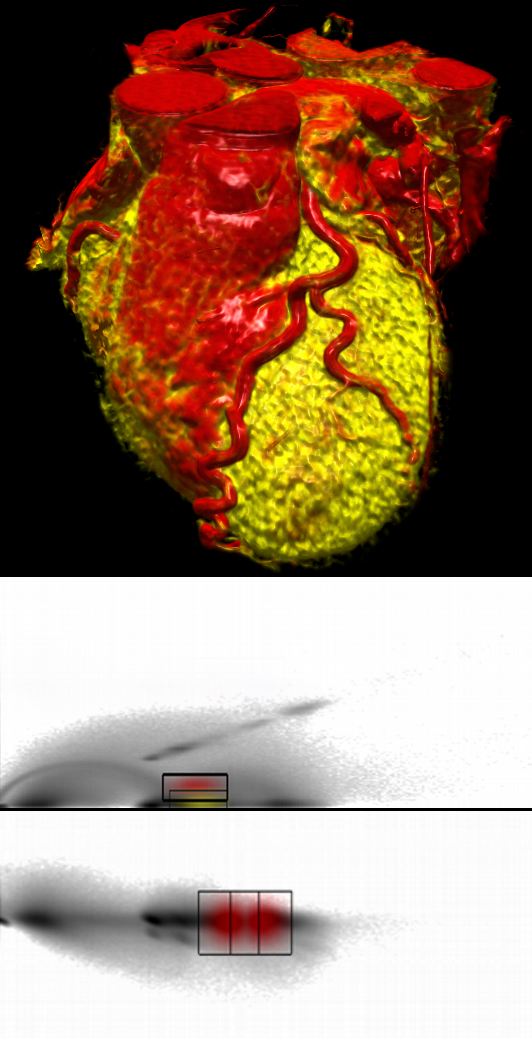
The software started as a basic volume renderer for medical data using hardware accelerated texture slicing and ray casting algorithms and basic one-dimensional classification methods. Later, the project grew into a plugin-based framework supporting different kinds of volume (DICOM, NRRD, raw) and mesh based data formats and therefore formed a basis for applying our research and integrating visualization techniques to improve the diagnosis of medical data. It provides 2D slice based views on the data as well as high quality 3D volume rendering, multi-dimensional classification techniques, interactive clipping, data segmentation and manual coregistration tools. While the software allows 2D and 3D visualization of different modalities like PET, CT or MRI, the focus is on the combined visualization of quantitative perfusion PET and CT angiography for the diagnosis of ischemia.
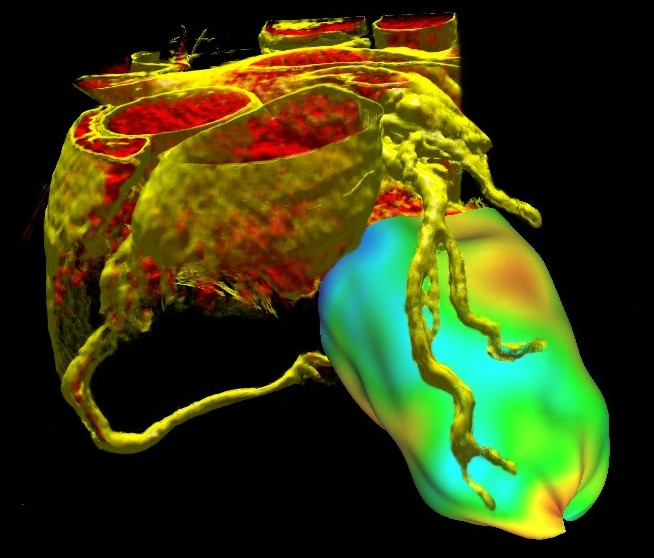
To put it simply, the aim of our work was to combine information from these two modalities in one visualization. In that way we want to give physicians a better overview as well as a detailed insight into the context of the data, thus improving the efficiency of the diagnosis. Examples are given in Figure 2 and 3. They show a combination of a CT scan and a quantitative PET perfusion of a patient's heart. In this case the focus was to analyze the coronary vessels for constrictions causing a low perfusion of the myocardial muscle. Therefore we used the CT scan to visualize the structure of the coronary vessels and added a visualization of the PET perfusion study (derived at the Heart and Diabetes Center). This information was mapped on a 3D object that resembles the mid layer of the left ventricle. The color mapping gives a clear overview of the level of perfusion in different regions of the muscle. The combination of both modalities shows very quickly, which arteries might cause problems simply by analyzing which vessels are suppling those areas of the heart, that are not well enough perfused (showing up in blueish colors). Performing the same diagnosis on the pictures independently - maybe even on 2D representations - would take essentially longer.
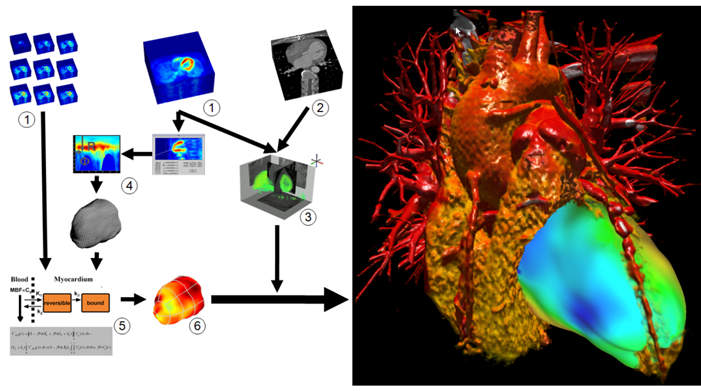
Figure 3 shows the data flow in detail: First, PET (1) and CT (2) currently are co-registered manually (3) (as shown in Figure 4). A PET uptake image (e.g. the last frame of a dynamic study using N-13-ammonia) is used for coregistration and for defining a surface inside the left myocardium (4). The blood flow at points on this surface is computed by compartment modeling (5). A converter transforms the functional data (6) derived from modeling back into anatomical space, where it can be added to the visualization. To highlight clinical relevant details from the CT study, i.e. the coronary arteries, without the interference of other anatomical structures, we use two dimensional transfer functions to assign specific visual attributes to the voxels of the features of interest. We are currently testing alternative transfer functions, such as texture-based and size-based functions.
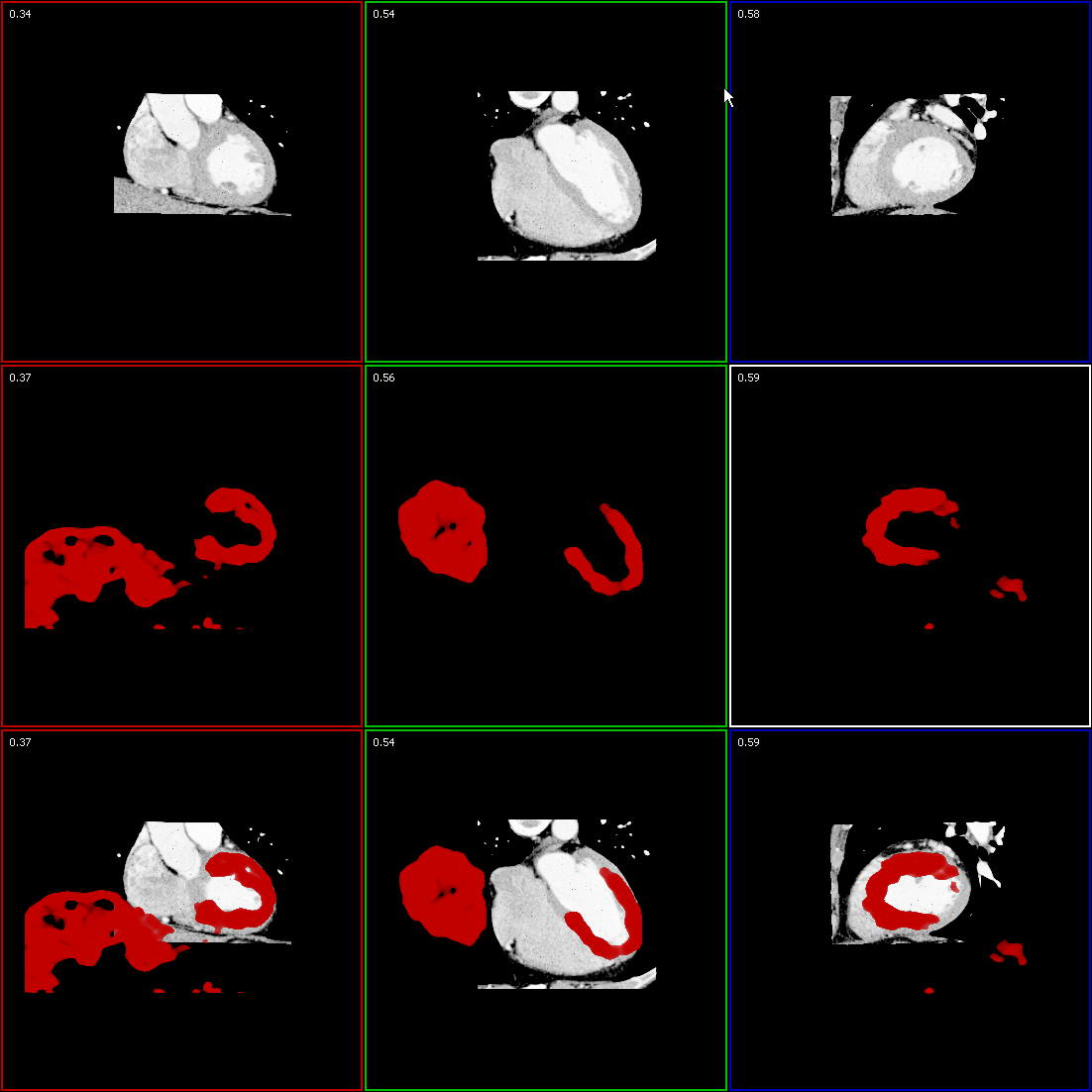
CT scans contain a combination of different materials and tissue types with overlapping boundaries. Depending on the actual diagnostic target, some of those features of the dataset must be highlighted, some must be shown transparent to retain the anatomical context while others should become completely invisible to prevent obscuring the more important parts. To extract the features of interest while suppressing insignificant parts, we developed a new approach for semi-automatic data classification that combines a fast semi-automatic classification to provide instant results with the flexibility of an histogram-based interactive approach. Due to variance in data of cardiac CT scans caused mainly by unequal distribution of contrast agent, gating and specific anatomy of the scanned subject, classification based on voxel’s value is not suitable. Instead, our method focuses on pattern detection in 2D joint histograms of data value and its first and second derivative. Although the histograms also change with data variation, neural networks are able to detect clusters and arcs in the histogram structure which represent features of interest that should be visualized. An example of a cardiac CT dataset after classification with the neural network using data value, gradient and second derivative is given in Figure 5.
3D - Data, Dimension, Diagnosis
Our work at an exhibition at the worlds largest computer museum
From 25th October 2006 - 20th May 2007 the special exhibition Computer.Medicine took place at the Heinz Nixdorf MuseumsForum (HNF) - the worlds largest computer museum. For this exhibition we prepared a special version of Volume Studio, running in a presentation mode, demonstrating the possibilities and advantages of computer technology and 3D visualizations in the medical area. The most intriguing feature of this demonstration is probably the stereo view we implemented especially for the exhibition. The software is presented on a big screen in a cinema-like room (Software Theater) using a stereo projection system. By using stereo glasses the audience may experience visualizations of medical data like a heart and even a whole body scan with the sensation of real 3D.
Publications
| [HZR+15] | Haas, N.A.; Zelle, M.; Rosendahl, W.; Zink, A.; Preuss, R.; Laser, T. K.; Gostner, R. P.; Arens, S.; Domik, G.; Burchert, W.: Hypoplastic left heart in the 6500-year-old Detmold Child, The Lancet, June 2015, Vol. 385, pp. 2432 |
| [ADW+14] | Arens, S.; Domik G.; Weise R.; Holzinger, J.; Burchert W.: Optimierte 3D-‐ Morphologiedarstellung tomographischer Datensätze des Herzens, 53. Jahrestagung der Deutschen Gesellschaft für Nuklearmedizin Hannover 2014 |
| [HZR+14] | Haas, N.A., M. Zelle, W. Rosendahl, A. Zink, R, P. Gostner, S. Arens, G. Domik, R. Preuss, T.K. Laser, W. Burchert, 2014, “A young heart from old ages – diagnosis of a hypoplastic left heart syndrome in the 6500 year old mummy ‘The Detmold Child’”, accepted for The Lancet. |
| [ABD13] | Arens S., Bolte M., Domik G.: Pull-based Dataflow for Multi-Volume Visualization. Tech. Rep. tr-ri-13-332, University of Paderborn, Depart- ment of Computer Science, June 2013. |
| [ABD11] | S. Arens, M. Bolte, G. Domik |
| [AD11] | S. Arens, G. Domik |
| [DAS+11] | G. Domik, S. Arens, I. Scharlau, F. Hilkenmeier |
| [AIB+11] | S. Arens, T. Isenberg, M. Bolte, G. Domik, R. Weise, J. Holzinger, H. Fricke, W. Burchert: "Räumliche Wahrnehmung ct-angiographischer Koronararterien in einer Curved Planar Reformation mittels Direct Volume Rendering". Tagungsband der Gemeinsamen Jahrestagung NuklearMedizin 2011 der Deutschen, Österreichischen und Schweizer Gesellschaften fuer Nuklearmedizin, Bregenz, Austria, 2011 |
| [AD10] | S. Arens, G. Domik: "A Survey of Transfer Functions Suitable for Volume Rendering". 8th IEEE/EG International Symposium on Volume Graphics, Norrköping, Sweden, 2010 |
| [ADW+10] | S. Arens, G. Domik, R. Weise, H. Fricke, W. Burchert: "Weniger Sehen – mehr Verstehen: Größen-basierte und Textur-basierte Transferfunktionen zur Volumenvisualisierung von CT Daten der Koronargefäße". Tagungsband der 41. Jahrestagung der Deutschen Gesellschaft für Medizinische Physik, Freiburg i. Br., Germany, 2010 |
| [WAE+10] | P. Wette, S. Arens, A. Elsner, G. Domik: Extending the Corkskrew Algorithms to Find Bifurcations of Vessels. Proceedings of IASTED Computer Graphics and Imaging, Innsbruck, Austria, 2010 |
| [FEW+09] | Fricke, H., Elsner, A., Weise, R., Bolte, M., van den Hoff, J., Burchert, W., Domik, G., Fricke, E., 2009, Quantitative myocardial perfusion PET combined with coronary anatomy derived from CT angiography: Validation of a new fusion and visualisation software, Zeitschrift für Medizinische Physik, Elsevier. |
| [EFF+09] | A. Elsner, E. Fricke, H. Fricke, G. Domik, R. Weise, W. Burchert. Evaluation of Manual Coregistration for Cardiac PET/CT from Independent Scanners. 47. Jahrestagung der Deutschen Gesellschaft für Nuklearmedizin, Leipzig, Germany, April 2009. |
| [ZE1+08] | D. Zukic, A. Elsner, Z. Avdagic, G. Domik. Neural networks in 3D medical scan visualization. In Int. Conf. on Computer Graphics and Artificial Intelligence (3IA 2008), Technological Educational Institute of Athens, May 2008. |
| [EFF+08] | A. Elsner, E. Fricke, H. Fricke, G. Domik, R. Weise, W. Burchert. Semiautomatische Klassifizierung von CT–Datensätzen zur kombinierten PET/CT-Darstellung. 46. Jahrestagung der Deutschen Gesellschaft für Nuklearmedizin, Leipzig, Germany, April 2008. |
| [ZEA+07] | Dženan Zukic, Andreas Elsner, Zikrija Avdagic, Gitta Domik, Wilhelm Schäfer, Dušanka Boškovic. Neural Networks in CT scan visualization. In Proc. of the 21st International Symposium on Information, Communication and Automation Technologies (ICAT 2007), Sarajevo, Bosnia Herzegovina, Faculty of Electrical Engineering, University of Sarajevo, October 2007. |
| [FWW+07] | H. Fricke, M. Schwier, R. Weise, A. Elsner, E. Fricke, G. Domik, W. Burchert. Coregistration and visualization of cardiac CT studies and dynamic PET studies using scene-graphs, direct volume rendering and 2D transfer functions. In: Proceedings of the 54th SNM Annual Meeting, Washington D.C., USA, May 2007. |
| [SDG+07] | Schwier M, Domik G, Goetz F, Fricke E, Weise R, Fricke H, Burchert W. Interaktive 2D-Transferfunktionen zur Visualisierung und Auswertung von CT/PET Datensätzen (Interactive 2D-Transfer Functions to Visualize and Evaluate CT/PET Data ), 45. Jahrestagung der Deutschen Gesellschaft für Nuklearmedizin; Hannover, Germany, 2007. |
| [Sch07] | |
| [BDF+07] | Burchert, W., Domik, G., Fricke, E., Fricke, H., Götz, F., and Weise, R., 2007, Neue Hoffnung für Herzkranke - Volumenrendering für die Gesundheit, ForschungsForum Paderborn (Titel und ab Seite 6), Ausgabe 2007, Universität Paderborn, Germany. |
| [LGW+06] | |
| [GJD05] | Goetz F, Junklewitz T, Domik G. Real-Time Marching Cubes on the Vertex Shader. In: Eurographics 2005; Dublin, Ireland. |
| [DK06] | |
| [Kut07] | Kutter O. Use of Real-Time Shader to Visualize Static and Dynamic Particles and Flows [Master Thesis]: University of Paderborn; 2006. |
| [KW06] | Koch, C. und Wiesemann, T., 2006, Development of a Framework with Graphical Editor to Visualize Medical Data [Bachelor Thesis]: University of Paderborn, 2006. |
Bachelor and Master Theses with Volume Studio
2014
2013
2012
2011
2010
2009
2008
2007